Search
- Page Path
- HOME > Search
- Isolation and identification of monkeypox virus MPXV-ROK-P1-2022 from the first case in the Republic of Korea
- Jin-Won Kim, Minji Lee, Hwachul Shin, Chi-Hwan Choi, Myung-Min Choi, Jee Woong Kim, Hwajung Yi, Cheon-Kwon Yoo, Gi-Eun Rhie
- Osong Public Health Res Perspect. 2022;13(4):308-311. Published online August 31, 2022
- DOI: https://doi.org/10.24171/j.phrp.2022.0232
- 5,307 View
- 136 Download
- 7 Web of Science
- 8 Crossref
-
 Graphical Abstract
Graphical Abstract
 Abstract
Abstract
 PDF
PDF 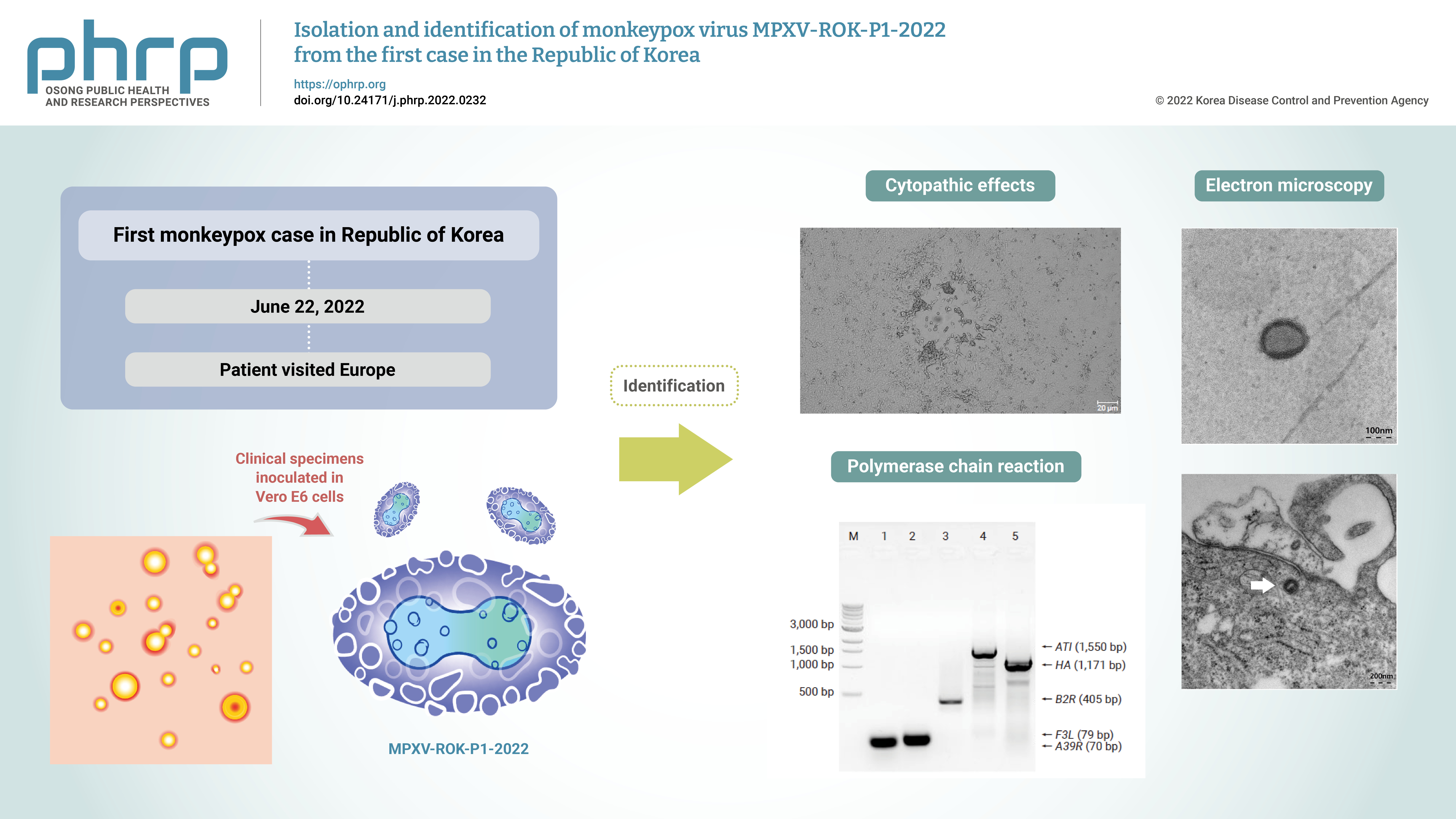
- Objectives
Monkeypox outbreaks in nonendemic countries have been reported since early May 2022. The first case of monkeypox in the Republic of Korea was confirmed in a patient who traveled to Europe in June 2022, and an attempt was made to isolate and identify the monkeypox virus (MPXV) from the patient’s specimens.
Methods
Clinical specimens from the patient were inoculated in Vero E6 cells. The isolated virus was identified as MPXV by the observation of cytopathic effects on Vero E6 cells, transmission electron microscopy, conventional polymerase chain reaction (PCR), and sequencing of PCR products.
Results
Cytopathic effects were observed in Vero E6 cells that were inoculated with skin lesion swab eluates. After multiple passages from the primary culture, orthopoxvirus morphology was observed using transmission electron microscopy. In addition, both MPXV-specific (F3L and ATI) and orthopoxvirus-specific genes (A39R, B2R, and HA) were confirmed by conventional PCR and Sanger sequencing.
Conclusion
These results indicate the successful isolation and identification of MPXV from the first patient in the Republic of Korea. The isolated virus was named MPXV-ROK-P1-2022. -
Citations
Citations to this article as recorded by- Ultrasensitive one-pot detection of monkeypox virus with RPA and CRISPR in a sucrose-aided multiphase aqueous system
Yue Wang, Yixin Tang, Yukang Chen, Guangxi Yu, Xue Zhang, Lihong Yang, Chenjie Zhao, Pei Wang, Song Gao, Frederick S. B. Kibenge, Ruijie Deng, Wei Chen, Shuang Yang
Microbiology Spectrum.2024;[Epub] CrossRef - Epidemiological, Clinical, and Virological Investigation of the First Four Cases of Monkeypox in Cartagena during the 2022 Outbreak
Steev Loyola, Mashiel Fernández-Ruiz, Doris Gómez-Camargo
Pathogens.2023; 12(2): 159. CrossRef - 원숭이두창바이러스의 분리 배양과 전장유전체 정보 분석
민지 이, 진원 김, 치환 최, 화철 신, 명민 최, 상은 이, 화중 이, 윤석 정
Public Health Weekly Report.2023; 16(15): 464. CrossRef - Overview of Diagnostic Methods, Disease Prevalence and Transmission of Mpox (Formerly Monkeypox) in Humans and Animal Reservoirs
Ravendra P. Chauhan, Ronen Fogel, Janice Limson
Microorganisms.2023; 11(5): 1186. CrossRef - How to cope with suspected mpox patients in the outpatient clinic
Nam Joong Kim, Sun Huh
Journal of the Korean Medical Association.2023; 66(5): 325. CrossRef - An Updated Review on Monkeypox Viral Disease: Emphasis on Genomic Diversity
Ali A. Rabaan, Nada A. Alasiri, Mohammed Aljeldah, Abeer N. Alshukairiis, Zainab AlMusa, Wadha A. Alfouzan, Abdulmonem A. Abuzaid, Aref A. Alamri, Hani M. Al-Afghani, Nadira Al-baghli, Nawal Alqahtani, Nadia Al-baghli, Mashahed Y. Almoutawa, Maha Mahmoud
Biomedicines.2023; 11(7): 1832. CrossRef - Monkeypox (Mpox) virus isolation and ultrastructural characterisation from a Brazilian human sample case
Milene Dias Miranda, Gabriela Cardoso Caldas, Vivian Neuza Ferreira, Ortrud Monika Barth, Aline de Paula Dias da Silva, Mayara Secco Torres Silva, Beatriz Grinsztejn, Valdiléa Gonçalves Veloso, Thiago Moreno Souza, Edson Elias da Silva, Debora Ferreira Ba
Memórias do Instituto Oswaldo Cruz.2023;[Epub] CrossRef - Isolation and Characterization of Monkeypox Virus from the First Case of Monkeypox — Chongqing Municipality, China, 2022
Baoying Huang, Hua Zhao, Jingdong Song, Li Zhao, Yao Deng, Wen Wang, Roujian Lu, Wenling Wang, Jiao Ren, Fei Ye, Houwen Tian, Guizhen Wu, Hua Ling, Wenjie Tan
China CDC Weekly.2022; 4(46): 1019. CrossRef
- Ultrasensitive one-pot detection of monkeypox virus with RPA and CRISPR in a sucrose-aided multiphase aqueous system
- Genomic Surveillance of SARS-CoV-2: Distribution of Clades in the Republic of Korea in 2020
- Ae Kyung Park, Il-Hwan Kim, Junyoung Kim, Jeong-Min Kim, Heui Man Kim, Chae young Lee, Myung-Guk Han, Gi-Eun Rhie, Donghyok Kwon, Jeong-Gu Nam, Young-Joon Park, Jin Gwack, Nam-Joo Lee, SangHee Woo, Jin Sun No, Jaehee Lee, Jeemin Ha, JeeEun Rhee, Cheon-Kwon Yoo, Eun-Jin Kim
- Osong Public Health Res Perspect. 2021;12(1):37-43. Published online February 23, 2021
- DOI: https://doi.org/10.24171/j.phrp.2021.12.1.06
- 9,166 View
- 223 Download
- 21 Web of Science
- 22 Crossref
-
 Abstract
Abstract
 PDF
PDF Since a novel beta-coronavirus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was first reported in December 2019, there has been a rapid global spread of the virus. Genomic surveillance was conducted on samples isolated from infected individuals to monitor the spread of genetic variants of SARS-CoV-2 in Korea. The Korea Disease Control and Prevention Agency performed whole genome sequencing of SARS-CoV-2 in Korea for 1 year (January 2020 to January 2021). A total of 2,488 SARS-CoV-2 cases were sequenced (including 648 cases from abroad). Initially, the prevalent clades of SARS-CoV-2 were the S and V clades, however, by March 2020, GH clade was the most dominant. Only international travelers were identified as having G or GR clades, and since the first variant 501Y.V1 was identified (from a traveler from the United Kingdom on December 22nd, 2020), a total of 27 variants of 501Y.V1, 501Y.V2, and 484K.V2 have been classified (as of January 25th, 2021). The results in this study indicated that quarantining of travelers entering Korea successfully prevented dissemination of the SARS-CoV-2 variants in Korea.
-
Citations
Citations to this article as recorded by- Increased viral load in patients infected with severe acute respiratory syndrome coronavirus 2 Omicron variant in the Republic of Korea
Jeong-Min Kim, Dongju Kim, Nam-Joo Lee, Sang Hee Woo, Jaehee Lee, Hyeokjin Lee, Ae Kyung Park, Jeong-Ah Kim, Chae Young Lee, Il-Hwan Kim, Cheon Kwon Yoo, Eun-Jin Kim
Osong Public Health and Research Perspectives.2023; 14(4): 272. CrossRef - Rapid Emergence of the Omicron Variant of Severe Acute Respiratory Syndrome Coronavirus 2 in Korea
Ae Kyung Park, Il-Hwan Kim, Chae Young Lee, Jeong-Ah Kim, Hyeokjin Lee, Heui Man Kim, Nam-Joo Lee, SangHee Woo, Jaehee Lee, JeeEun Rhee, Cheon-Kwon Yoo, Eun-Jin Kim
Annals of Laboratory Medicine.2023; 43(2): 211. CrossRef - A Seroprevalence Study on Residents in a Senior Care Facility with Breakthrough SARS-CoV-2 Omicron Infection
Heui Man Kim, Eun Ju Lee, Sang Won O, Yong Jun Choi, Hyeokjin Lee, Sae Jin Oh, Jeong-Min Kim, Ae Kyung Park, Jeong-Ah Kim, Chae young Lee, Jong Mu Kim, Hanul Park, Young Joon Park, Jeong-Hee Yu, Eun-Young Kim, Hwa-Pyeong Ko, Eun-Jin Kim
Viral Immunology.2023; 36(3): 203. CrossRef - COVID-19 Cases and Deaths among Healthcare Personnel with the Progression of the Pandemic in Korea from March 2020 to February 2022
Yeonju Kim, Sung-Chan Yang, Jinhwa Jang, Shin Young Park, Seong Sun Kim, Chansoo Kim, Donghyok Kwon, Sang-Won Lee
Tropical Medicine and Infectious Disease.2023; 8(6): 308. CrossRef - The COVID-19 pandemic and healthcare utilization in Iran: evidence from an interrupted time series analysis
Monireh Mahmoodpour-Azari, Satar Rezaei, Nasim Badiee, Mohammad Hajizadeh, Ali Mohammadi, Ali Kazemi-Karyani, Shahin Soltani, Mehdi Khezeli
Osong Public Health and Research Perspectives.2023; 14(3): 180. CrossRef - Online Phylogenetics with matOptimize Produces Equivalent Trees and is Dramatically More Efficient for Large SARS-CoV-2 Phylogenies than de novo and Maximum-Likelihood Implementations
Alexander M Kramer, Bryan Thornlow, Cheng Ye, Nicola De Maio, Jakob McBroome, Angie S Hinrichs, Robert Lanfear, Yatish Turakhia, Russell Corbett-Detig, Olivier Gascuel
Systematic Biology.2023; 72(5): 1039. CrossRef - Genomic epidemiology of SARS-CoV-2 variants in South Korea between January 2020 and February 2023
Il-Hwan Kim, Jin Sun No, Jeong-Ah Kim, Ae Kyung Park, HyeokJin Lee, Jeong-Min Kim, Nam-Joo Lee, Chi-Kyeong Kim, Chae Young Lee, SangHee Woo, Jaehee Lee, JeeEun Rhee, Eun-Jin Kim
Virology.2023; 587: 109869. CrossRef - Genomic evidence of SARS‐CoV‐2 reinfection in the Republic of Korea
Ae Kyung Park, Jee Eun Rhee, Il‐Hwan Kim, Heui Man Kim, Hyeokjin Lee, Jeong‐Ah Kim, Chae Young Lee, Nam‐Joo Lee, SangHee Woo, Jaehee Lee, Jin Sun No, Gi‐Eun Rhie, Seong Jin Wang, Sang‐Eun Lee, Young Joon Park, Gemma Park, Jung Yeon Kim, Jin Gwack, Cheon‐K
Journal of Medical Virology.2022; 94(4): 1717. CrossRef - SARS-CoV-2 B.1.619 and B.1.620 Lineages, South Korea, 2021
Ae Kyung Park, Il-Hwan Kim, Heui Man Kim, Hyeokjin Lee, Nam-Joo Lee, Jeong-Ah Kim, SangHee Woo, Chae young Lee, Jaehee Lee, Sae Jin Oh, JeeEun Rhee, Cheon-Kwon Yoo, Eun-Jin Kim
Emerging Infectious Diseases.2022; 28(2): 415. CrossRef - Humoral and Cellular Responses to COVID-19 Vaccines in SARS-CoV-2 Infection-Naïve and -Recovered Korean Individuals
Ji-Young Hwang, Yunhwa Kim, Kyung-Min Lee, Eun-Jeong Jang, Chang-Hoon Woo, Chang-Ui Hong, Seok-Tae Choi, Sivilay Xayaheuang, Jong-Geol Jang, June-Hong Ahn, Hosun Park
Vaccines.2022; 10(2): 332. CrossRef - Increase in Viral Load in Patients With SARS-CoV-2 Delta Variant Infection in the Republic of Korea
Jeong-Min Kim, Jee Eun Rhee, Myeongsu Yoo, Heui Man Kim, Nam-Joo Lee, Sang Hee Woo, Hye-Jun Jo, Donghyok Kwon, Sangwon Lee, Cheon Kwon Yoo, Eun-Jin Kim
Frontiers in Microbiology.2022;[Epub] CrossRef - Molecular Dynamics Studies on the Structural Stability Prediction of SARS-CoV-2 Variants Including Multiple Mutants
Kwang-Eun Choi, Jeong-Min Kim, Jee Eun Rhee, Ae Kyung Park, Eun-Jin Kim, Cheon Kwon Yoo, Nam Sook Kang
International Journal of Molecular Sciences.2022; 23(9): 4956. CrossRef - SARS-CoV-2 shedding dynamics and transmission in immunosuppressed patients
Jee-Soo Lee, Ki Wook Yun, Hyeonju Jeong, Boram Kim, Man Jin Kim, Jae Hyeon Park, Ho Seob Shin, Hyeon Sae Oh, Hobin Sung, Myung Gi Song, Sung Im Cho, So Yeon Kim, Chang Kyung Kang, Pyoeng Gyun Choe, Wan Beom Park, Nam Joong Kim, Myoung-Don Oh, Eun Hwa Choi
Virulence.2022; 13(1): 1242. CrossRef - Immunological and Pathological Peculiarity of Severe Acute Respiratory Syndrome Coronavirus 2 Beta Variant
Sunhee Lee, Gun Young Yoon, Su Jin Lee, Young-Chan Kwon, Hyun Woo Moon, Yu-Jin Kim, Haesoo Kim, Wooseong Lee, Gi Uk Jeong, Chonsaeng Kim, Kyun-Do Kim, Seong-Jun Kim, Dae-Gyun Ahn, Miguel Angel Martinez
Microbiology Spectrum.2022;[Epub] CrossRef - Clinical scoring system to predict viable viral shedding in patients with COVID-19
Sung Woon Kang, Heedo Park, Ji Yeun Kim, Sunghee Park, So Yun Lim, Sohyun Lee, Joon-Yong Bae, Jeonghun Kim, Seongman Bae, Jiwon Jung, Min Jae Kim, Yong Pil Chong, Sang-Oh Lee, Sang-Ho Choi, Yang Soo Kim, Sung-Cheol Yun, Man-Seong Park, Sung-Han Kim
Journal of Clinical Virology.2022; 157: 105319. CrossRef - Model-informed COVID-19 exit strategy with projections of SARS-CoV-2 infections generated by variants in the Republic of Korea
Sung-mok Jung, Kyungmin Huh, Munkhzul Radnaabaatar, Jaehun Jung
BMC Public Health.2022;[Epub] CrossRef - Comparative analysis of mutational hotspots in the spike protein of SARS-CoV-2 isolates from different geographic origins
Sanghoo Lee, Mi-Kyeong Lee, Hyeongkyun Na, Jinwoo Ahn, Gayeon Hong, Youngkee Lee, Jimyeong Park, Yejin Kim, Yun-Tae Kim, Chang-Ki Kim, Hwan-Sub Lim, Kyoung-Ryul Lee
Gene Reports.2021; 23: 101100. CrossRef - Review of Current COVID-19 Diagnostics and Opportunities for Further Development
Yan Mardian, Herman Kosasih, Muhammad Karyana, Aaron Neal, Chuen-Yen Lau
Frontiers in Medicine.2021;[Epub] CrossRef - Locally harvested Covid-19 convalescent plasma could probably help combat the geographically determined SARS-CoV-2 viral variants
Manish Raturi, Anuradha Kusum, Mansi Kala, Garima Mittal, Anita Sharma, Naveen Bansal
Transfusion Clinique et Biologique.2021; 28(3): 300. CrossRef - Molecular Dynamics Studies on the Structural Characteristics for the Stability Prediction of SARS-CoV-2
Kwang-Eun Choi, Jeong-Min Kim, JeeEun Rhee, Ae Kyung Park, Eun-Jin Kim, Nam Sook Kang
International Journal of Molecular Sciences.2021; 22(16): 8714. CrossRef - Management following the first confirmed case of SARS-CoV-2 in a domestic cat associated with a massive outbreak in South Korea
Taewon Han, Boyeong Ryu, Suyeon Lee, Yugyeong Song, Yoongje Jeong, Ilhwan Kim, Jeongmin Kim, Eunjin Kim, Wonjun Lee, Hyunju Lee, Haekyoung Hwang
One Health.2021; 13: 100328. CrossRef - Genomic epidemiology reveals the reduction of the introduction and spread of SARS-CoV-2 after implementing control strategies in Republic of Korea, 2020
Jung-Hoon Kwon, Jeong-Min Kim, Dong-hun Lee, Ae Kyung Park, Il-Hwan Kim, Da-Won Kim, Ji-Yun Kim, Noori Lim, Kyeong-Yeon Cho, Heui Man Kim, Nam-Joo Lee, SangHee Woo, Chae Young Lee, Jin Sun No, Junyoung Kim, JeeEun Rhee, Myung-Guk Han, Gi-Eun Rhie, Cheon K
Virus Evolution.2021;[Epub] CrossRef
- Increased viral load in patients infected with severe acute respiratory syndrome coronavirus 2 Omicron variant in the Republic of Korea
- 2019 Tabletop Exercise for Laboratory Diagnosis and Analyses of Unknown Disease Outbreaks by the Korea Centers for Disease Control and Prevention
- Il-Hwan Kim, Jun Hyeong Jang, Su-Kyoung Jo, Jin Sun No, Seung-Hee Seo, Jun-Young Kim, Sang-Oun Jung, Jeong-Min Kim, Sang-Eun Lee, Hye-Kyung Park, Eun-Jin Kim, Jun Ho Jeon, Myung-Min Choi, Boyeong Ryu, Yoon Suk Jang, Hwami Kim, Jin Lee, Seung-Hwan Shin, Hee Kyoung Kim, Eun-Kyoung Kim, Ye Eun Park, Cheon-Kwon Yoo, Sang-Won Lee, Myung-Guk Han, Gi-Eun Rhie, Byung Hak Kang
- Osong Public Health Res Perspect. 2020;11(5):280-285. Published online October 22, 2020
- DOI: https://doi.org/10.24171/j.phrp.2020.11.5.03
- 5,869 View
- 106 Download
-
 Abstract
Abstract
 PDF
PDF Objectives The Korea Centers for Disease Control and Prevention has published “A Guideline for Unknown Disease Outbreaks (UDO).” The aim of this report was to introduce tabletop exercises (TTX) to prepare for UDO in the future.
Methods The UDO Laboratory Analyses Task Force in Korea Centers for Disease Control and Prevention in April 2018, assigned unknown diseases into 5 syndromes, designed an algorithm for diagnosis, and made a panel list for diagnosis by exclusion. Using the guidelines and laboratory analyses for UDO, TTX were introduced.
Results Since September 9th, 2018, the UDO Laboratory Analyses Task Force has been preparing TTX based on a scenario of an outbreak caused by a novel coronavirus. In December 2019, through TTX, individual missions, epidemiological investigations, sample treatments, diagnosis by exclusions, and next generation sequencing analysis were discussed, and a novel coronavirus was identified as the causal pathogen.
Conclusion Guideline and laboratory analyses for UDO successfully applied in TTX. Conclusions drawn from TTX could be applied effectively in the analyses for the initial response to COVID-19, an ongoing epidemic of 2019 – 2020. Therefore, TTX should continuously be conducted for the response and preparation against UDO.
- Genome-Wide Identification and Characterization of Point Mutations in the SARS-CoV-2 Genome
- Jun-Sub Kim, Jun-Hyeong Jang, Jeong-Min Kim, Yoon-Seok Chung, Cheon-Kwon Yoo, Myung-Guk Han
- Osong Public Health Res Perspect. 2020;11(3):101-111. Published online May 14, 2020
- DOI: https://doi.org/10.24171/j.phrp.2020.11.3.05
- 15,367 View
- 518 Download
- 89 Web of Science
- 64 Crossref
-
 Abstract
Abstract
 PDF
PDF Objectives Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged in Wuhan, China, in December 2019 and has been rapidly spreading worldwide. Although the causal relationship among mutations and the features of SARS-CoV-2 such as rapid transmission, pathogenicity, and tropism, remains unclear, our results of genomic mutations in SARS-CoV-2 may help to interpret the interaction between genomic characterization in SARS-CoV-2 and infectivity with the host.
Methods A total of 4,254 genomic sequences of SARS-CoV-2 were collected from the Global Initiative on Sharing all Influenza Data (GISAID). Multiple sequence alignment for phylogenetic analysis and comparative genomic approach for mutation analysis were conducted using Molecular Evolutionary Genetics Analysis (MEGA), and an in-house program based on Perl language, respectively.
Results Phylogenetic analysis of SARS-CoV-2 strains indicated that there were 3 major clades including S, V, and G, and 2 subclades (G.1 and G.2). There were 767 types of synonymous and 1,352 types of non-synonymous mutation. ORF1a, ORF1b, S, and N genes were detected at high frequency, whereas ORF7b and E genes exhibited low frequency. In the receptor-binding domain (RBD) of the S gene, 11 non-synonymous mutations were observed in the region adjacent to the angiotensin converting enzyme 2 (ACE2) binding site.
Conclusion It has been reported that the rapid infectivity and transmission of SARS-CoV-2 associated with host receptor affinity are derived from several mutations in its genes. Without these genetic mutations to enhance evolutionary adaptation, species recognition, host receptor affinity, and pathogenicity, it would not survive. It is expected that our results could provide an important clue in understanding the genomic characteristics of SARS-CoV-2.
-
Citations
Citations to this article as recorded by- Three-Dimensional Structural Stability and Local Electrostatic Potential at Point Mutations in Spike Protein of SARS-CoV-2 Coronavirus
Svetlana H. Hristova, Alexandar M. Zhivkov
International Journal of Molecular Sciences.2024; 25(4): 2174. CrossRef - SARS-CoV-2-ORF3a variant Q57H reduces its pro-apoptotic activity in host cells
Maria Landherr, Iuliia Polina, Michael W. Cypress, Isabel Chaput, Bridget Nieto, Bong Sook Jhun, Jin O-Uchi
F1000Research.2024; 13: 331. CrossRef - Geographical distribution of host's specific SARS-CoV-2 mutations in the early phase of the COVID-19 pandemic
Mohammad Khalid, David Murphy, Maryam Shoai, Jonahunnatha Nesson George-William, Yousef Al-ebini
Gene.2023; 851: 147020. CrossRef - SARS-CoV-2 mutations on diagnostic gene targets in the second wave in Zimbabwe: A retrospective genomic analysis
C Nyagupe, L de Oliveira Martins, H Gumbo, T Mashe, T Takawira, KK Maeka, A Juru, LK Chikanda, AR Tauya, AJ Page, RA Kingsley, R Simbi, J Chirenda, J Manasa, V Ruhanya, RT Mavenyengwa
South African Medical Journal.2023; 113(3): 141. CrossRef - A low dose of RBD and TLR7/8 agonist displayed on influenza virosome particles protects rhesus macaque against SARS-CoV-2 challenge
Gerrit Koopman, Mario Amacker, Toon Stegmann, Ernst J. Verschoor, Babs E. Verstrepen, Farien Bhoelan, Denzel Bemelman, Kinga P. Böszörményi, Zahra Fagrouch, Gwendoline Kiemenyi-Kayere, Daniella Mortier, Dagmar E. Verel, Henk Niphuis, Roja Fidel Acar, Ivan
Scientific Reports.2023;[Epub] CrossRef - Detection of IgA and IgG Antibodies against the Structural Proteins of SARS-CoV-2 in Breast Milk and Serum Samples Derived from Breastfeeding Mothers
Karen Cortés-Sarabia, Vianey Guzman-Silva, Karla Montserrat Martinez-Pacheco, Jesús Alberto Meza-Hernández, Víctor Manuel Luna-Pineda, Marco Antonio Leyva-Vázquez, Amalia Vences-Velázquez, Fredy Omar Beltrán-Anaya, Oscar Del Moral-Hernández, Berenice Illa
Viruses.2023; 15(4): 966. CrossRef - Evaluation of antiviral drugs against newly emerged SARS-CoV-2 Omicron subvariants
Junhyung Cho, Younmin Shin, Jeong-Sun Yang, Jun Won Kim, Kyung-Chang Kim, Joo-Yeon Lee
Antiviral Research.2023; 214: 105609. CrossRef - Genetic Analysis and Epitope Prediction of SARS-CoV-2 Genome in Bahia, Brazil: An In Silico Analysis of First and Second Wave Genomics Diversity
Gabriela Andrade, Guilherme Matias, Lara Chrisóstomo, João da Costa-Neto, Juan Sampaio, Arthur Silva, Isaac Cansanção
COVID.2023; 3(5): 655. CrossRef - Natural selection shapes the evolution of SARS-CoV-2 Omicron in Bangladesh
Mohammad Tanbir Habib, Saikt Rahman, Mokibul Hassan Afrad, Arif Mahmud Howlader, Manjur Hossain Khan, Farhana Khanam, Ahmed Nawsher Alam, Emran Kabir Chowdhury, Ziaur Rahman, Mustafizur Rahman, Tahmina Shirin, Firdausi Qadri
Frontiers in Genetics.2023;[Epub] CrossRef - Unraveling the impact of ORF3a Q57H mutation on SARS-CoV-2: insights from molecular dynamics
Md. Jahirul Islam, Md. Siddik Alom, Md. Shahadat Hossain, Md Ackas Ali, Shaila Akter, Shafiqul Islam, M. Obayed Ullah, Mohammad A. Halim
Journal of Biomolecular Structure and Dynamics.2023; : 1. CrossRef - Genomic characterization of SARS-CoV-2 from Uganda using MinION nanopore sequencing
Praiscillia Kia, Eric Katagirya, Fredrick Elishama Kakembo, Doreen Ato Adera, Moses Luutu Nsubuga, Fahim Yiga, Sharley Melissa Aloyo, Brendah Ronah Aujat, Denis Foe Anguyo, Fred Ashaba Katabazi, Edgar Kigozi, Moses L. Joloba, David Patrick Kateete
Scientific Reports.2023;[Epub] CrossRef - Molecular definition of severe acute respiratory syndrome coronavirus 2 receptor‐binding domain mutations: Receptor affinity versus neutralization of receptor interaction
Monique Vogel, Gilles Augusto, Xinyue Chang, Xuelan Liu, Daniel Speiser, Mona O. Mohsen, Martin F. Bachmann
Allergy.2022; 77(1): 143. CrossRef - Peptides and peptidomimetics as therapeutic agents for Covid‐19
Achyut Dahal, Jafrin Jobayer Sonju, Konstantin G. Kousoulas, Seetharama D. Jois
Peptide Science.2022;[Epub] CrossRef - Emergence of unique SARS-CoV-2 ORF10 variants and their impact on protein structure and function
Sk. Sarif Hassan, Kenneth Lundstrom, Ángel Serrano-Aroca, Parise Adadi, Alaa A.A. Aljabali, Elrashdy M. Redwan, Amos Lal, Ramesh Kandimalla, Tarek Mohamed Abd El-Aziz, Pabitra Pal Choudhury, Gajendra Kumar Azad, Samendra P. Sherchan, Gaurav Chauhan, Murta
International Journal of Biological Macromolecules.2022; 194: 128. CrossRef - COVID-19: comprehensive review on mutations and current vaccines
Ananda Vardhan Hebbani, Swetha Pulakuntla, Padmavathi Pannuru, Sreelatha Aramgam, Kameswara Rao Badri, Vaddi Damodara Reddy
Archives of Microbiology.2022;[Epub] CrossRef - Prediction of the Effects of Nonsynonymous Variants on SARS-CoV-2 Proteins
Boon Zhan Sia, Wan Xin Boon, Yoke Yee Yap, Shalini Kumar, Chong Han Ng
F1000Research.2022; 11: 9. CrossRef - Identification of SARS-CoV-2 Variants and Their Clinical Significance in Hefei, China
Xiao-wen Cheng, Jie Li, Lu Zhang, Wen-jun Hu, Lu Zong, Xiang Xu, Jin-ping Qiao, Mei-juan Zheng, Xi-wen Jiang, Zhi-kun Liang, Yi-fan Zhou, Ning Zhang, Hua-qing Zhu, Yuan-hong Xu
Frontiers in Medicine.2022;[Epub] CrossRef - Promising inhibitors against main protease of SARS CoV-2 from medicinal plants: In silico identification
OLUWAKEMI EBENEZER, MICHAEL SHAPI
Acta Pharmaceutica.2022; 72(2): 159. CrossRef - Genomic surveillance of SARS-CoV-2 in the state of Delaware reveals tremendous genomic diversity
Karl R. Franke, Robert Isett, Alan Robbins, Carrie Paquette-Straub, Craig A. Shapiro, Mary M. Lee, Erin L. Crowgey, Pierre Roques
PLOS ONE.2022; 17(1): e0262573. CrossRef - The importance of accessory protein variants in the pathogenicity of SARS-CoV-2
Sk. Sarif Hassan, Pabitra Pal Choudhury, Guy W. Dayhoff, Alaa A.A. Aljabali, Bruce D. Uhal, Kenneth Lundstrom, Nima Rezaei, Damiano Pizzol, Parise Adadi, Amos Lal, Antonio Soares, Tarek Mohamed Abd El-Aziz, Adam M. Brufsky, Gajendra Kumar Azad, Samendra P
Archives of Biochemistry and Biophysics.2022; 717: 109124. CrossRef - The mutational dynamics of the SARS-CoV-2 virus in serial passages in vitro
Sissy Therese Sonnleitner, Stefanie Sonnleitner, Eva Hinterbichler, Hannah Halbfurter, Dominik B.C. Kopecky, Stephan Koblmüller, Christian Sturmbauer, Wilfried Posch, Gernot Walder
Virologica Sinica.2022; 37(2): 198. CrossRef - Evolutionary dynamics of SARS-CoV-2 circulating in Yogyakarta and Central Java, Indonesia: sequence analysis covering furin cleavage site (FCS) region of the spike protein
Nastiti Wijayanti, Faris Muhammad Gazali, Endah Supriyati, Mohamad Saifudin Hakim, Eggi Arguni, Marselinus Edwin Widyanto Daniwijaya, Titik Nuryastuti, Matin Nuhamunada, Rahma Nabilla, Sofia Mubarika Haryana, Tri Wibawa
International Microbiology.2022; 25(3): 531. CrossRef - Prediction of the effects of the top 10 nonsynonymous variants from 30229 SARS-CoV-2 strains on their proteins
Boon Zhan Sia, Wan Xin Boon, Yoke Yee Yap, Shalini Kumar, Chong Han Ng
F1000Research.2022; 11: 9. CrossRef - The Mutational Landscape of SARS-CoV-2 Variants of Concern Recovered From Egyptian Patients in 2021
Mohamed G. Seadawy, Reem Binsuwaidan, Badriyah Alotaibi, Thanaa A. El-Masry, Bassem E. El-Harty, Ahmed F. Gad, Walid F. Elkhatib, Maisra M. El-Bouseary
Frontiers in Microbiology.2022;[Epub] CrossRef - Genomic surveillance, evolution and global transmission of SARS-CoV-2 during 2019–2022
Nadim Sharif, Khalid J. Alzahrani, Shamsun Nahar Ahmed, Afsana Khan, Hamsa Jameel Banjer, Fuad M. Alzahrani, Anowar Khasru Parvez, Shuvra Kanti Dey, Jayanta Bhattacharya
PLOS ONE.2022; 17(8): e0271074. CrossRef - Comparison of Intracellular Transcriptional Response of NHBE Cells to Infection with SARS-CoV-2 Washington and New York Strains
Tiana M. Scott, Antonio Solis-Leal, J. Brandon Lopez, Richard A. Robison, Bradford K. Berges, Brett E. Pickett
Frontiers in Cellular and Infection Microbiology.2022;[Epub] CrossRef - The Delta and Omicron Variants of SARS-CoV-2: What We Know So Far
Vivek Chavda, Rajashri Bezbaruah, Kangkan Deka, Lawandashisha Nongrang, Tutumoni Kalita
Vaccines.2022; 10(11): 1926. CrossRef - SARS-CoV-2’NİN SÜREGELEN EVRİMİ: PANDEMİNİN SONUNA NE KADAR YAKINIZ?
Elmas Pınar KAHRAMAN KILBAŞ, Mustafa ALTINDİŞ
Journal of Biotechnology and Strategic Health Rese.2022; 6(3): 201. CrossRef - Complete Genomic Characterisation and Mutation Patterns of Iraqi SARS-CoV-2 Isolates
Jivan Qasim Ahmed, Sazan Qadir Maulud
Diagnostics.2022; 13(1): 8. CrossRef - Comprehensive annotations of the mutational spectra of SARS‐CoV‐2 spike protein: a fast and accurate pipeline
Mohammad Shaminur Rahman, Mohammad Rafiul Islam, Mohammad Nazmul Hoque, Abu Sayed Mohammad Rubayet Ul Alam, Masuda Akther, Joynob Akter Puspo, Salma Akter, Azraf Anwar, Munawar Sultana, Mohammad Anwar Hossain
Transboundary and Emerging Diseases.2021; 68(3): 1625. CrossRef - Genomic and proteomic mutation landscapes of SARS‐CoV‐2
Christian Luke D. C. Badua, Karol Ann T. Baldo, Paul Mark B. Medina
Journal of Medical Virology.2021; 93(3): 1702. CrossRef - Variant analysis of the first Lebanese SARS-CoV-2 isolates
Mhamad Abou-Hamdan, Kassem Hamze, Ali Abdel Sater, Haidar Akl, Nabil El-zein, Israa Dandache, Fadi Abdel-sater
Genomics.2021; 113(1): 892. CrossRef - Global SNP analysis of 11,183 SARS‐CoV‐2 strains reveals high genetic diversity
Fangfeng Yuan, Liping Wang, Ying Fang, Leyi Wang
Transboundary and Emerging Diseases.2021; 68(6): 3288. CrossRef - SARS-CoV-2 hot-spot mutations are significantly enriched within inverted repeats and CpG island loci
Pratik Goswami, Martin Bartas, Matej Lexa, Natália Bohálová, Adriana Volná, Jiří Červeň, Veronika Červeňová, Petr Pečinka, Vladimír Špunda, Miroslav Fojta, Václav Brázda
Briefings in Bioinformatics.2021; 22(2): 1338. CrossRef - Deciphering the Subtype Differentiation History of SARS-CoV-2 Based on a New Breadth-First Searching Optimized Alignment Method Over a Global Data Set of 24,768 Sequences
Qianyu Lin, Yunchuanxiang Huang, Ziyi Jiang, Feng Wu, Lan Ma
Frontiers in Genetics.2021;[Epub] CrossRef - Temporal increase in D614G mutation of SARS-CoV-2 in the Middle East and North Africa
Malik Sallam, Nidaa A. Ababneh, Deema Dababseh, Faris G. Bakri, Azmi Mahafzah
Heliyon.2021; 7(1): e06035. CrossRef - Environmental aspect and applications of nanotechnology to eliminate COVID-19 epidemiology risk
Eman Serag, Marwa El-Zeftawy
Nanotechnology for Environmental Engineering.2021;[Epub] CrossRef - Genomic Characterization and Phylogenetic Analysis of SARS-CoV-2 in Libya
Silvia Fillo, Francesco Giordani, Anella Monte, Giovanni Faggioni, Riccardo De Santis, Nino D’Amore, Stefano Palomba, Taher Hamdani, Kamel Taloa, Atef Belkhir Jumaa, Siraj Bitrou, Ahmed Alaruusi, Wadie Mad, Abdulaziz Zorgani, Omar Elahmer, Badereddin Anna
Microbiology Research.2021; 12(1): 138. CrossRef - SARS-CoV-2 Entry Related Viral and Host Genetic Variations: Implications on COVID-19 Severity, Immune Escape, and Infectivity
Szu-Wei Huang, Sheng-Fan Wang
International Journal of Molecular Sciences.2021; 22(6): 3060. CrossRef - Emergence of novel SARS-CoV-2 variants in the Netherlands
Aysun Urhan, Thomas Abeel
Scientific Reports.2021;[Epub] CrossRef - Lung organoid simulations for modelling and predicting the effect of mutations on SARS-CoV-2 infectivity
Sally Esmail, Wayne R. Danter
Computational and Structural Biotechnology Journal.2021; 19: 1701. CrossRef - MSC-derived exosomes carrying a cocktail of exogenous interfering RNAs an unprecedented therapy in era of COVID-19 outbreak
Monire Jamalkhah, Yasaman Asaadi, Mohammadreza Azangou-Khyavy, Javad Khanali, Masoud Soleimani, Jafar Kiani, Ehsan Arefian
Journal of Translational Medicine.2021;[Epub] CrossRef - Mutations and Epidemiology of SARS-CoV-2 Compared to Selected Corona Viruses during the First Six Months of the COVID-19 Pandemic: A Review
Mirriam M. Nzivo, Nancy L.M. Budambula
Journal of Pure and Applied Microbiology.2021; 15(2): 524. CrossRef - The First Molecular Characterization of Serbian SARS-CoV-2 Isolates From a Unique Early Second Wave in Europe
Danijela Miljanovic, Ognjen Milicevic, Ana Loncar, Dzihan Abazovic, Dragana Despot, Ana Banko
Frontiers in Microbiology.2021;[Epub] CrossRef - Identification of a High-Frequency Intrahost SARS-CoV-2 Spike Variant with Enhanced Cytopathic and Fusogenic Effects
Lynda Rocheleau, Geneviève Laroche, Kathy Fu, Corina M. Stewart, Abdulhamid O. Mohamud, Marceline Côté, Patrick M. Giguère, Marc-André Langlois, Martin Pelchat, Dimitrios Paraskevis
mBio.2021;[Epub] CrossRef - Notable and Emerging Variants of SARS-CoV-2 Virus: A Quick Glance
Sagar Dholariya, Deepak Narayan Parchwani, Ragini Singh, Amit Sonagra, Anita Motiani, Digishaben Patel
Indian Journal of Clinical Biochemistry.2021; 36(4): 451. CrossRef - Genomic characterization of SARS‐CoV‐2 isolates from patients in Turkey reveals the presence of novel mutations in spike and nsp12 proteins
Erdem Sahin, Gulendam Bozdayi, Selin Yigit, Hager Muftah, Murat Dizbay, Ozlem G. Tunccan, Isil Fidan, Kayhan Caglar
Journal of Medical Virology.2021; 93(10): 6016. CrossRef - Temporal landscape of mutational frequencies in SARS-CoV-2 genomes of Bangladesh: possible implications from the ongoing outbreak in Bangladesh
Otun Saha, Israt Islam, Rokaiya Nurani Shatadru, Nadira Naznin Rakhi, Md. Shahadat Hossain, Md. Mizanur Rahaman
Virus Genes.2021; 57(5): 413. CrossRef - SARS-CoV-2 receptor-binding mutations and antibody contact sites
Marios Mejdani, Kiandokht Haddadi, Chester Pham, Radhakrishnan Mahadevan
Antibody Therapeutics.2021; 4(3): 149. CrossRef - Evolutionary trajectory of SARS-CoV-2 and emerging variants
Jalen Singh, Pranav Pandit, Andrew G. McArthur, Arinjay Banerjee, Karen Mossman
Virology Journal.2021;[Epub] CrossRef - Molecular characterization of SARS-CoV-2 from Bangladesh: implications in genetic diversity, possible origin of the virus, and functional significance of the mutations
Md. Marufur Rahman, Shirmin Bintay Kader, S.M. Shahriar Rizvi
Heliyon.2021; 7(8): e07866. CrossRef - Exploring the Binding Mechanism of PF-07321332 SARS-CoV-2 Protease Inhibitor through Molecular Dynamics and Binding Free Energy Simulations
Bilal Ahmad, Maria Batool, Qurat ul Ain, Moon Suk Kim, Sangdun Choi
International Journal of Molecular Sciences.2021; 22(17): 9124. CrossRef - Characterization of the SARS-CoV-2 genomes in Egypt in first and second waves of infection
Abdel-Rahman N. Zekri, Abeer A. Bahnasy, Mohamed M. Hafez, Zeinab K. Hassan, Ola S. Ahmed, Hany K. Soliman, Enas R. El-Sisi, Mona H. Salah El Dine, May S. Solimane, Lamyaa S. Abdel Latife, Mohamed G. Seadawy, Ahmed S. Elsafty, Mohamed Abouelhoda
Scientific Reports.2021;[Epub] CrossRef - A Global Mutational Profile of SARS-CoV-2: A Systematic Review and Meta-Analysis of 368,316 COVID-19 Patients
Wardah Yusof, Ahmad Adebayo Irekeola, Yusuf Wada, Engku Nur Syafirah Engku Abd Rahman, Naveed Ahmed, Nurfadhlina Musa, Muhammad Fazli Khalid, Zaidah Abdul Rahman, Rosline Hassan, Nik Yusnoraini Yusof, Chan Yean Yean
Life.2021; 11(11): 1224. CrossRef - Phylogenetic and full-length genome mutation analysis of SARS-CoV-2 in Indonesia prior to COVID-19 vaccination program in 2021
Reviany V. Nidom, Setyarina Indrasari, Irine Normalina, Astria N. Nidom, Balqis Afifah, Lestari Dewi, Andra K. Putra, Arif N. M. Ansori, Muhammad K. J. Kusala, Mohammad Y. Alamudi, Chairul A. Nidom
Bulletin of the National Research Centre.2021;[Epub] CrossRef - Global Pandemic as a Result of Severe Acute Respiratory Syndrome Coronavirus 2 Outbreak: A Biomedical Perspective
Charles Arvind Sethuraman Vairavan, Devarani Rameshnathan, Nagaraja Suryadevara, Gnanendra Shanmugam
Journal of Pure and Applied Microbiology.2021; 15(4): 1759. CrossRef - Predicting the Molecular Mechanism of Sini Jia Renshen Decoction in Treating Severe COVID-19 Patients Based on Network Pharmacology and Molecular Docking
Yi Wen Liu, Ai Xia Yang, Li Lu, Tie Hua Huang
Natural Product Communications.2021; 16(12): 1934578X2110592. CrossRef - Can SARS-CoV-2 Accumulate Mutations in the S-Protein to Increase Pathogenicity?
Aditya K. Padhi, Timir Tripathi
ACS Pharmacology & Translational Science.2020; 3(5): 1023. CrossRef - Optimized Pseudotyping Conditions for the SARS-COV-2 Spike Glycoprotein
Marc C. Johnson, Terri D. Lyddon, Reinier Suarez, Braxton Salcedo, Mary LePique, Maddie Graham, Clifton Ricana, Carolyn Robinson, Detlef G. Ritter, Viviana Simon
Journal of Virology.2020;[Epub] CrossRef - A Crowned Killer’s Résumé: Genome, Structure, Receptors, and Origin of SARS-CoV-2
Shichuan Wang, Mirko Trilling, Kathrin Sutter, Ulf Dittmer, Mengji Lu, Xin Zheng, Dongliang Yang, Jia Liu
Virologica Sinica.2020; 35(6): 673. CrossRef - Host or pathogen-related factors in COVID-19 severity?
Christian Gortázar, Francisco J Rodríguez del-Río, Lucas Domínguez, José de la Fuente
The Lancet.2020; 396(10260): 1396. CrossRef - Host or pathogen-related factors in COVID-19 severity? – Authors' reply
Lucy C Okell, Robert Verity, Aris Katzourakis, Erik M Volz, Oliver J Watson, Swapnil Mishra, Patrick Walker, Charlie Whittaker, Christl A Donnelly, Steven Riley, Azra C Ghani, Axel Gandy, Seth Flaxman, Neil M Ferguson, Samir Bhatt
The Lancet.2020; 396(10260): 1397. CrossRef - Recent updates on COVID-19: A holistic review
Shweta Jakhmola, Omkar Indari, Dharmendra Kashyap, Nidhi Varshney, Annu Rani, Charu Sonkar, Budhadev Baral, Sayantani Chatterjee, Ayan Das, Rajesh Kumar, Hem Chandra Jha
Heliyon.2020; 6(12): e05706. CrossRef - Impact of Genetic Variability in ACE2 Expression on the Evolutionary Dynamics of SARS-CoV-2 Spike D614G Mutation
Szu-Wei Huang, Sorin O. Miller, Chia-Hung Yen, Sheng-Fan Wang
Genes.2020; 12(1): 16. CrossRef
- Three-Dimensional Structural Stability and Local Electrostatic Potential at Point Mutations in Spike Protein of SARS-CoV-2 Coronavirus
- Emergence of Norovirus GII.17-associated Outbreak and Sporadic Cases in Korea from 2014 to 2015
- Sunyoung Jung, Bo-Mi Hwang, HyunJu Jung, GyungTae Chung, Cheon-Kwon Yoo, Deog-Yong Lee
- Osong Public Health Res Perspect. 2017;8(1):86-90. Published online February 28, 2017
- DOI: https://doi.org/10.24171/j.phrp.2017.8.1.12
- 3,991 View
- 27 Download
- 8 Crossref
-
 Abstract
Abstract
 PDF
PDF Human norovirus are major causative agent of nonbacterial acute gastroenteritis. In general, genogroup (G) II.4 is the most prominent major genotype that circulate in human population and the environment. However, a shift in genotypic trends was observed in Korea in December 2014. In this study, we investigated the trend of norovirus genotype in detail using the database of Acute Diarrhea Laboratory Surveillance (K-EnterNet) in Korea. GII.17 has since become a major contributor to outbreaks of norovirus-related infections and sporadic cases in Korea, although the reason for this shift remain unknown.
-
Citations
Citations to this article as recorded by- Monitoring of foodborne viruses in pre- and post-washed root vegetables in the Republic of Korea
Sunho Park, Md Iqbal Hossain, Soontag Jung, Zhaoqi Wang, Daseul Yeo, Seoyoung Woo, Yeeun Seo, Myeong-In Jeong, Changsun Choi
Food Control.2023; 154: 109982. CrossRef - Epidemiology of Norovirus Outbreaks Reported to the Public Health Emergency Event Surveillance System, China, 2014–2017
Yiyao Lian, Shuyu Wu, Li Luo, Bin Lv, Qiaohong Liao, Zhongjie Li, Jeanette J. Rainey, Aron J. Hall, Lu Ran
Viruses.2019; 11(4): 342. CrossRef - Nearly Complete Genome Sequence of a Human Norovirus GII.P17-GII.17 Strain Isolated from Brazil in 2015
Cristina Santiso-Bellón, Azahara Fuentes-Trillo, Juliana da Silva Ribeiro de Andrade, Carolina Monzó, Susana Vila-Vicent, Roberto Gozalbo Rovira, Javier Buesa, Felipe J. Chaves, Marize Pereira Miagostovich, Jesús Rodríguez-Díaz, Jelle Matthijnssens
Microbiology Resource Announcements.2019;[Epub] CrossRef - Development of SNP Markers for Norovirus Related FUT2 Gene in Oyster
守泉 闫
Hans Journal of Biomedicine.2019; 09(03): 135. CrossRef - Research on the contamination levels of norovirus in food facilities using groundwater in South Korea, 2015–2016
Jeong Su Lee, In Sun Joo, Si Yeon Ju, Min Hee Jeong, Yun-Hee Song, Hyo Sun Kwak
International Journal of Food Microbiology.2018; 280: 35. CrossRef - Norovirus GII.17 Associated with a Foodborne Acute Gastroenteritis Outbreak in Brazil, 2016
Juliana da Silva Ribeiro de Andrade, Tulio Machado Fumian, José Paulo Gagliardi Leite, Matheus Ribeiro de Assis, Alexandre Madi Fialho, Sergio Mouta, Cristiane Mendes Pereira Santiago, Marize Pereira Miagostovich
Food and Environmental Virology.2018; 10(2): 212. CrossRef - Phylogenetic characterization of norovirus strains detected from sporadic gastroenteritis in Seoul during 2014–2016
Young Eun Kim, Miok Song, Jaein Lee, Hyun Jung Seung, Eun-Young Kwon, Jinkyung Yu, Youngok Hwang, Taeho Yoon, Tae Jun Park, In Kyoung Lim
Gut Pathogens.2018;[Epub] CrossRef - The prevalence of non-GII.4 norovirus genotypes in acute gastroenteritis outbreaks in Jinan, China
Lanzheng Liu, Hengyun Guan, Ying Zhang, Chunrong Wang, Guoliang Yang, Shiman Ruan, Huailong Zhao, Xiuyun Han, Adriana Calderaro
PLOS ONE.2018; 13(12): e0209245. CrossRef
- Monitoring of foodborne viruses in pre- and post-washed root vegetables in the Republic of Korea
- Occurrence of Norovirus GII.4 Sydney Variant-related Outbreaks in Korea
- Sunyoung Jung, Bo-Mi Hwang, Hyun Ju Jeong, Gyung Tae Chung, Cheon-Kwon Yoo, Yeon-Ho Kang, Deog-Yong Lee
- Osong Public Health Res Perspect. 2015;6(5):322-326. Published online October 31, 2015
- DOI: https://doi.org/10.1016/j.phrp.2015.10.004
- 2,900 View
- 17 Download
- 8 Crossref
-
 Abstract
Abstract
 PDF
PDF - Human noroviruses are major causative agents of food and waterborne outbreaks of nonbacterial acute gastroenteritis. In this study, we report the epidemiological features of three outbreak cases of norovirus in Korea, and we describe the clinical symptoms and distribution of the causative genotypes. The incidence rates of the three outbreaks were 16.24% (326/2,007), 4.1% (27/656), and 16.8% (36/214), respectively. The patients in these three outbreaks were affected by acute gastroenteritis. These schools were provided unheated food from the same manufacturing company. Two genotypes (GII.3 and GII.4) of the norovirus were detected in these cases. Among them, major causative strains of GII.4 (Hu-jeju-47-2007KR-like) were identified in patients, food handlers, and groundwater from the manufacturing company of the unheated food. In the GII.4 (Hu-jeju-47-2007KR-like) strain of the norovirus, the nucleotide sequences were identical and identified as the GII.4 Sydney variant. Our data suggests that the combined epidemiological and laboratory results were closely related, and the causative pathogen was the GII.4 Sydney variant strain from contaminated groundwater.
-
Citations
Citations to this article as recorded by- A systematic review and meta-analysis indicates a substantial burden of human noroviruses in shellfish worldwide, with GII.4 and GII.2 being the predominant genotypes
Yijing Li, Liang Xue, Junshan Gao, Weicheng Cai, Zilei Zhang, Luobing Meng, Shuidi Miao, Xiaojing Hong, Mingfang Xu, Qingping Wu, Jumei Zhang
Food Microbiology.2023; 109: 104140. CrossRef - Molecular epidemiology of norovirus infections in children with acute gastroenteritis in 2017–2019 in Tianjin, China
Yulian Fang, Yanzhi Zhang, Hong Wang, Ouyan Shi, Wei Wang, Mengzhu Hou, Lu Wang, Jinying Wu, Yu Zhao
Journal of Medical Virology.2022; 94(2): 616. CrossRef - Assessment of potential infectivity of human norovirus in the traditional Korean salted clam product “Jogaejeotgal” by floating electrode-dielectric barrier discharge plasma
Eun Bi Jeon, Man-Seok Choi, Ji Yoon Kim, Eun Ha Choi, Jun Sup Lim, Jinsung Choi, Kwang Soo Ha, Ji Young Kwon, Sang Hyeon Jeong, Shin Young Park
Food Research International.2021; 141: 110107. CrossRef - Characterizing the effects of thermal treatment on human norovirus GII.4 viability using propidium monoazide combined with RT-qPCR and quality assessments in mussels
Eun Bi Jeon, Man-Seok Choi, Ji Yoon Kim, Kwang Soo Ha, Ji Young Kwon, Sung Hyeon Jeong, Hee Jung Lee, Yeoun Joong Jung, Ji-Hyoung Ha, Shin Young Park
Food Control.2020; 109: 106954. CrossRef - Molecular epidemiology of genogroup II norovirus infections in acute gastroenteritis patients during 2014–2016 in Pudong New Area, Shanghai, China
Caoyi Xue, Lifeng Pan, Weiping Zhu, Yuanping Wang, Huiqin Fu, Chang Cui, Lan Lu, Sun Qiao, Biao Xu
Gut Pathogens.2018;[Epub] CrossRef - Review: Epidemiological evidence of groundwater contribution to global enteric disease, 1948–2015
Heather M. Murphy, Morgan D. Prioleau, Mark A. Borchardt, Paul D. Hynds
Hydrogeology Journal.2017; 25(4): 981. CrossRef - Change in Concentrations of Human Norovirus and Male-Specific Coliphage under Various Temperatures, Salinities, and pH Levels in Seawater
Poong Ho Kim, Yong Soo Park, Kunbawui Park, Ji Young Kwon, Hong Sik Yu, Hee Jung Lee, Ji Hoe Kim, Tae Seek Lee
Korean Journal of Fisheries and Aquatic Sciences.2016; 49(4): 454. CrossRef - Norovirus outbreaks occurred in different settings in the Republic of Korea
Hae-Wol Cho, Chaeshin Chu
Osong Public Health and Research Perspectives.2015; 6(5): 281. CrossRef
- A systematic review and meta-analysis indicates a substantial burden of human noroviruses in shellfish worldwide, with GII.4 and GII.2 being the predominant genotypes
- Epidemics of Norovirus GII.4 Variant in Outbreak Cases in Korea, 2004–2012
- Sunyoung Jung, Hyun Ju Jeong, Bo-Mi Hwang, Cheon-Kwon Yoo, Gyung Tae Chung, Hyesook Jeong, Yeon-Ho Kang, Deog-Yong Lee
- Osong Public Health Res Perspect. 2015;6(5):318-321. Published online October 31, 2015
- DOI: https://doi.org/10.1016/j.phrp.2015.10.002
- 3,013 View
- 20 Download
- 2 Crossref
-
 Abstract
Abstract
 PDF
PDF - Norovirus GII.4 is recognized as a worldwide cause of nonbacterial outbreaks. In particular, the GII.4 variant occurs every 2–3 years according to antigenic variation. The aim of our study was to identify GII.4 variants in outbreaks in Korea during 2004–2012. Partial VP1 sequence of norovirus GII.4-related outbreaks during 2004–2012 was analyzed. The partial VP1 sequence was detected with reverse transcription-polymerase chain reaction, seminested polymerase chain reaction, and nucleotide sequence of 312-314 base pairs for phylogenetic comparison. Nine variants emerged in outbreaks, with the Sydney variant showing predominance recently. This predominance may persist for at least 3 years, although new variants may appear in Korea.
-
Citations
Citations to this article as recorded by- Trends for Syndromic Surveillance of Norovirus in Emergency Department Data Based on Chief Complaints
Soyeoun Kim, Sohee Kim, Bo Youl Choi, Boyoung Park
The Journal of Infectious Diseases.2023;[Epub] CrossRef - Genotypic and Epidemiological Trends of Acute Gastroenteritis Associated with Noroviruses in China from 2006 to 2016
Shu-Wen Qin, Ta-Chien Chan, Jian Cai, Na Zhao, Zi-Ping Miao, Yi-Juan Chen, She-Lan Liu
International Journal of Environmental Research an.2017; 14(11): 1341. CrossRef
- Trends for Syndromic Surveillance of Norovirus in Emergency Department Data Based on Chief Complaints
- Enteric Bacteria Isolated from Diarrheal Patients in Korea in 2014
- Nan-Ok Kim, Su-Mi Jung, Hae-Young Na, Gyung Tae Chung, Cheon-Kwon Yoo, Won Keun Seong, Sahyun Hong
- Osong Public Health Res Perspect. 2015;6(4):233-240. Published online August 31, 2015
- DOI: https://doi.org/10.1016/j.phrp.2015.07.005
- 3,131 View
- 22 Download
- 15 Crossref
-
 Abstract
Abstract
 PDF
PDF Supplementary Material
Supplementary Material - Objectives
The aim of this study was to characterize the pathogens responsible for causing diarrhea according to season, region of isolation, patient age, and sex as well as to provide useful data for the prevention of diarrheal disease.
Methods
Stool specimens from 14,886 patients with diarrhea were collected to identify pathogenic bacteria from January 2014 to December 2014 in Korea. A total of 3,526 pathogenic bacteria were isolated and analyzed according to season, region of isolation, and the age and sex of the patient.
Results
The breakdown of the isolated pathogenic bacteria were as follows: Salmonella spp. 476 (13.5%), pathogenic Escherichia coli 777 (22.0%), Vibrio parahaemolyticus 26 (0.74%), Shigella spp. 13 (0.37%), Campylobacter spp. 215 (6.10%), Clostridium perfringens 508 (14.4%), Staphylococcus aureus 1,144 (32.4%), Bacillus cereus 356 (10.1%), Listeria monocytogenes 1 (0.03%), and Yersinia enterocolitica 10 (0.3%). The isolation rate trend showed the highest ratio in the summer season from June to September for most of the pathogenic bacteria except the Gram-positive bacteria. The isolation rate of most of the pathogenic bacteria by patient age showed highest ratio in the 0–19 year age range. For isolation rate by region, 56.2% were isolated from cities and 43.8% were isolated from provinces.
Conclusion
Hygiene education should be addressed for diarrheal disease-susceptible groups, such as those younger than 10 years, aged 10–19 years, and older than 70 years, and monitoring for the pathogens is still required. In addition, an efficient laboratory surveillance system for infection control should be continued. -
Citations
Citations to this article as recorded by- Antimicrobial Susceptibility Patterns and Genetic Diversity of Campylobacter spp. Isolates from Patients with Diarrhea in South Korea
So Yeon Kim, Dongheui An, Hyemi Jeong, Jonghyun Kim
Microorganisms.2024; 12(1): 94. CrossRef - Comparative evaluation of two molecular multiplex syndromic panels with acute gastroenteritis
Kuenyoul Park, Bo-Moon Shin
Diagnostic Microbiology and Infectious Disease.2024; 109(1): 116211. CrossRef - Antimicrobial Resistance Profiles and Molecular Characteristics of Salmonella enterica Serovar Agona Isolated from Food-Producing Animals During 2010–2020 in South Korea
Hyun-Ju Song, Sekendar Ali, Bo-Youn Moon, Hee Young Kang, Eun Jeong Noh, Tae-Sun Kim, Su-Jeong Kim, Ji-In Kim, Yun Jin Lee, Soon-Seek Yoon, Suk-Kyung Lim
Foodborne Pathogens and Disease.2024;[Epub] CrossRef - An Outbreak of Campylobacter Jejuni Involving Healthcare Workers Detected by COVID-19 Healthcare Worker Symptom Surveillance
Hye Jin Shi, Jae Baek Lee, Shinhee Hong, Joong Sik Eom, Yoonseon Park
Korean Journal of Healthcare-Associated Infection .2023; 28(1): 172. CrossRef - Plant-derived nanoparticles as alternative therapy against Diarrheal pathogens in the era of antimicrobial resistance: A review
Tesleem Olatunde Abolarinwa, Daniel Jesuwenu Ajose, Bukola Opeyemi Oluwarinde, Justine Fri, Kotsoana Peter Montso, Omolola Esther Fayemi, Adeyemi Oladapo Aremu, Collins Njie Ateba
Frontiers in Microbiology.2022;[Epub] CrossRef - Prevalence, co-infection and seasonality of fecal enteropathogens from diarrheic cats in the Republic of Korea (2016–2019): a retrospective study
Ye-In Oh, Kyoung-Won Seo, Do-Hyung Kim, Doo-Sung Cheon
BMC Veterinary Research.2021;[Epub] CrossRef - Genotyping and antimicrobial susceptibility of Clostridium perfringens isolated from dromedary camels, pastures and herders
Mahmoud Fayez, Ibrahim Elsohaby, Theeb Al-Marri, Kamal Zidan, Ali Aldoweriej, Elham El-Sergany, Ahmed Elmoslemany
Comparative Immunology, Microbiology and Infectiou.2020; 70: 101460. CrossRef - Comparative Analysis of Aerotolerance, Antibiotic Resistance, and Virulence Gene Prevalence in Campylobacter jejuni Isolates from Retail Raw Chicken and Duck Meat in South Korea
Jinshil Kim, Hyeeun Park, Junhyung Kim, Jong Hyun Kim, Jae In Jung, Seongbeom Cho, Sangryeol Ryu, Byeonghwa Jeon
Microorganisms.2019; 7(10): 433. CrossRef - Guideline for the Antibiotic Use in Acute Gastroenteritis
Youn Jeong Kim, Ki-Ho Park, Dong-Ah Park, Joonhong Park, Byoung Wook Bang, Seung Soon Lee, Eun Jung Lee, Hyo-Jin Lee, Sung Kwan Hong, Yang Ree Kim
Infection & Chemotherapy.2019; 51(2): 217. CrossRef - Winning the War against Multi-Drug Resistant Diarrhoeagenic Bacteria
Chizoba Mercy Enemchukwu, Angus Nnamdi Oli, Ebere Innocent Okoye, Nonye Treasure Ujam, Emmanuel O. Osazuwa, George Ogonna Emechebe, Kenneth Nchekwube Okeke, Christian Chukwuemeka Ifezulike, Obiora Shedrack Ejiofor, Jude Nnaemeka Okoyeh
Microorganisms.2019; 7(7): 197. CrossRef - Molecular Characterization of Enterotoxigenic Escherichia coli in Foodborne Outbreak
Sang-Hun Park, Hyun-Jung Seung, Hyo-Won Jeong, So-Yun Park, Ji-Hun Jung, Young-Hee Jin, Sung-Hee Han, Hee-Soon Kim, Jin-Seok Kim, Joo-Hyun Park, Ye-Ji Gong, Chae Kyu Hong, Jib-Ho Lee, Il-Young Kim, Kweon Jung
Journal of Bacteriology and Virology.2018; 48(4): 113. CrossRef - Epidemiology of Listeria monocytogenes prevalence in foods, animals and human origin from Iran: a systematic review and meta-analysis
Reza Ranjbar, Mehrdad Halaji
BMC Public Health.2018;[Epub] CrossRef - The Epidemiological Influence of Climatic Factors on Shigellosis Incidence Rates in Korea
Yeong-Jun Song, Hae-Kwan Cheong, Myung Ki, Ji-Yeon Shin, Seung-sik Hwang, Mira Park, Moran Ki, Jiseun Lim
International Journal of Environmental Research an.2018; 15(10): 2209. CrossRef - Molecular characterization and antimicrobial resistance profile of Clostridium perfringens type A isolates from humans, animals, fish and their environment
Jay Prakash Yadav, Suresh Chandra Das, Pankaj Dhaka, Deepthi Vijay, Manesh Kumar, Asish Kumar Mukhopadhyay, Goutam Chowdhury, Pranav Chauhan, Rahul Singh, Kuldeep Dhama, Satya Veer Singh Malik, Ashok Kumar
Anaerobe.2017; 47: 120. CrossRef - Molecular characterization of antimicrobial resistant non-typhoidal Salmonella from poultry industries in Korea
Jin Eui Kim, Young ju Lee
Irish Veterinary Journal.2017;[Epub] CrossRef
- Antimicrobial Susceptibility Patterns and Genetic Diversity of Campylobacter spp. Isolates from Patients with Diarrhea in South Korea



 First
First Prev
Prev


